Intro
Previous CAct qPCRs from 20231210 (run 1 and run 2) exhibited lack of amplification in 100pg and 10pg input DNA quantities, as well as poor replication in 1000pg DNA input quantity, so I decided to see if poor replication was potentially related to poor sensitivity of the CAct primer (i.e. input DNA quantity was too low for accurate replication) set by running a 10,000pg (10ng) input DNA quantity.
qPCRs
All qPCRs were run in duplicate for each dilution (10,000pg, 1000pg, 100pg), with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in 20uL reactions. The CAct primers were utilized with an initial “working stock” concentration of 10uM and at a final concentration of 0.25uM in each qPCR reaction.
Unlike previous reactions, replicates were only performed in duplicate, as opposed to triplicate, due to limits in the quantity of the 10,000pg (10ng) stock. This stock was consists of pooled DNA (Notebook) from other samples and was instended to be able to serve as the source of positive control DNA for all subsequent qPCRs for this project. I didn’t want to use too much at this point in time and have the possibility of having to make a fresh stock and use even more of the individual DNA samples which still need to be used for the actual project.
qPCR calculations
- 20231210-qPCR_calcs (Google Sheet)
Results
Output files
Raw qPCR data (.pcrd; Requires CFX Maestro):
sam_2023-12-10_09-26-24_Connect.pcrd“Analysis Mode” was set to “Target” (“Fluorophore” is the default.)
Baseline threshold was set to “Regression”
qPCR data (CSV):
qPCR Report (PDF):
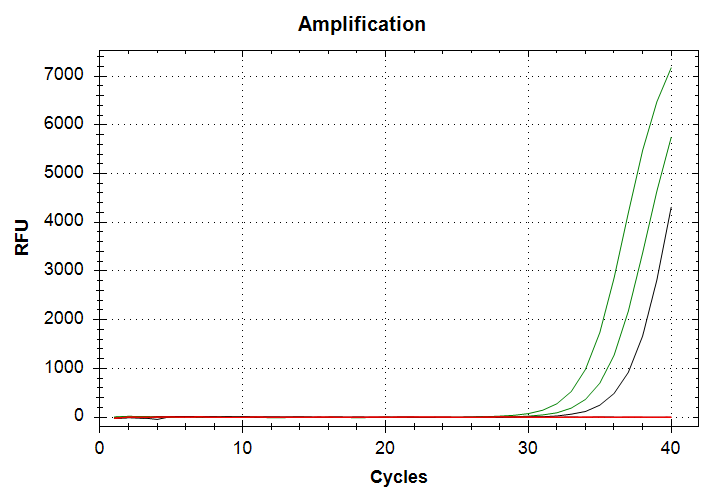
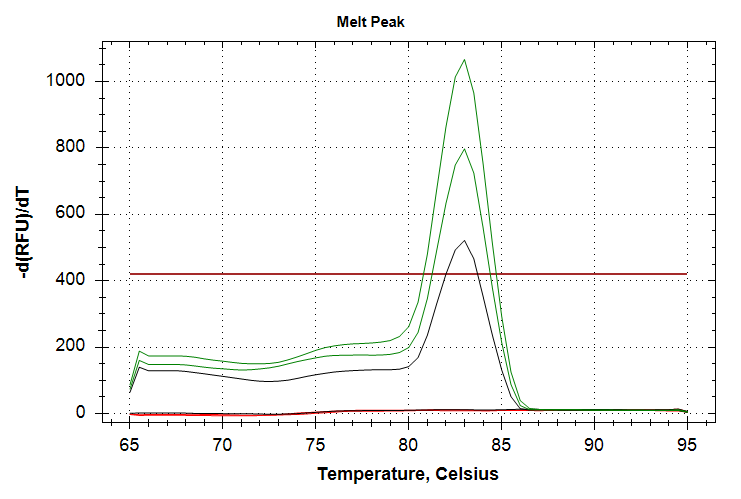
In the amplification plot, green lines are 10,000pg input DNA and correspond to Cq values of ~33 and ~35. Black line is 1,000pg input DNA and corresponds to a Cq value of ~37. We see both replicates of the 10,000pg input DNA samples produce amplificaiton, however the variation between replicates was attrocious. While with the 1,000pg input DNA sample, I’ve managed to reproduce this behavior across three different qPCRs! Only a single replicate ampflifies at a Cq of ~37. The resulting melt curves looks good (i.e. narrow peaks, same melt temp across reps/samples.).
Summary
At this point, I’m becoming suspicious of the input DNA, since the CAct primers are brand new, as well as the qPCR reagents. Additionally, a bigger concern is that the sensitivity (and/or the amount of C symbiont DNA present in samples) is too low to accurately detect across samples. Most of the samples are so dilute and/or have so little DNA that we will be unable to use 1,000pg of DNA per qPCR reaction - which is, essentially, the minimum quantity of DNA needed to get any detectable amplificaiton of the C symbiont DNA. Not entirely sure what the implications will be for the remainder of this project.